New Computational Method Enhances Protein Design Software
University of California (UC) San Francisco researchers, with support from the Synthetic Biology Engineering Research Center (Synberc), an NSF-funded center headquartered at UC Berkeley, collaborated with colleagues from global science company, DSM, to develop a new computational protein design method that drastically improves software used in computational biology. This advance greatly enhances the ability to use computer-aided design to accurately re-engineer the proteins that perform a vast array of functions within living organisms.
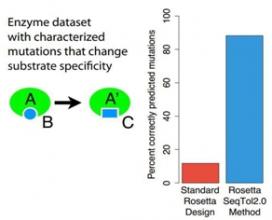
Computational protein design has advanced rapidly over the past decade. Despite many impressive successes, generating novel, functional proteins with activity levels similar to natural proteins remains challenging.The new computational protein design method helps researchers overcome this hurdle. It has been implemented in Rosetta, the state-of-the-art protein design software, resulting in a 75 percent increase in correct predictions. Rosetta is freely available to academic users and has been licensed by more than 20 companies.
Synberc’s mission centers on advancing biological solutions to real-world problems in health, energy, and the environment. Strategies to reengineer protein parts have increasing importance for expanding the range of accessible biological functions, such as the production of industrially valuable compounds, the detection and degradation of harmful substances, or the control of devices in response to small-molecule signals. The new method resulting from this Synberc-sponsored research is now actively used in several engineering applications within Synberc, at DSM, and beyond, and should be generally useful for many applications involving the re-design or optimization of protein part interaction specificity where a crystal structure of the protein is available.