New System Advances Genome Engineering for Medical Applications
Outcome/Accomplishment
Researchers at the Harvard and Columbia University Medical Schools have developed a novel system that enables more cost-effective and larger-scale genome engineering for a number of applications in synthetic biology and metabolic engineering. This research was supported by the NSF-funded Synthetic Biology Engineering Research Center (Synberc), which is headquartered at the University of California (UC) Berkeley, and in which Harvard and Columbia are partner institutions.
Impact/Benefits
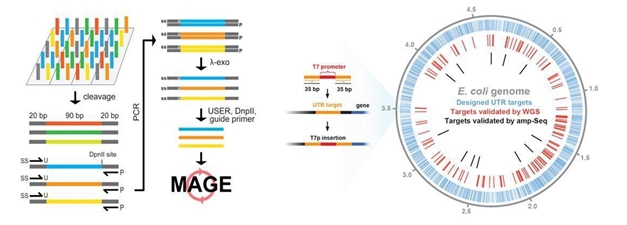
Genome engineering refers to strategies and techniques for targeted, specific modification of the genetic information (genome) of living organisms. Mutagenesis is a process by which the genome is changed in a stable manner, resulting in a mutation. Site-directed mutagenesis is a powerful tool in genome engineering—and the new Synberc system makes it more cost-effective and able to be performed on a larger scale that is more efficient and effective then existing methods.
Explanation/Background
Most traditional mutagenesis methods rely on the mutation of only one site at a time and efficiencies drop drastically when more than three sites are targeted simultaneously. The Synberc system combines Harvard’s Multiplex Automated Genome Engineering (MAGE) and Columbia’s Microarray Oligonucleotide (MO), to create MO-MAGE. MAGE allows simultaneous mutagenesis of multiple target sites in bacterial genomes using short oligonucleotides (such as the RNA or DNA molecules used in genetic research). However, this requires hundreds to thousands of unique oligos, which are costly to synthesize and impossible to scale up by traditional approaches. MO was developed to address the problem—it increases the number of oligos for direct use in altering thousands of genomic sites simultaneously.
One of the key milestones of synthetic biology is the development of “libraries” of diverse, well-characterized biological components that can be assembled to form new systems. The researchers’ vision for the MO-MAGE method is that it will be used to make thousands of specific chromosomal changes predicted to result in a desired phenotype (i.e., observable traits such height, eye color, and blood type) and combined with selection or screening of the cell library for promising mutants.
Outcome/Accomplishment
Researchers at the Harvard and Columbia University Medical Schools have developed a novel system that enables more cost-effective and larger-scale genome engineering for a number of applications in synthetic biology and metabolic engineering. This research was supported by the NSF-funded Synthetic Biology Engineering Research Center (Synberc), which is headquartered at the University of California (UC) Berkeley, and in which Harvard and Columbia are partner institutions.
Impact/benefits
Genome engineering refers to strategies and techniques for targeted, specific modification of the genetic information (genome) of living organisms. Mutagenesis is a process by which the genome is changed in a stable manner, resulting in a mutation. Site-directed mutagenesis is a powerful tool in genome engineering—and the new Synberc system makes it more cost-effective and able to be performed on a larger scale that is more efficient and effective then existing methods.
Explanation/Background
Most traditional mutagenesis methods rely on the mutation of only one site at a time and efficiencies drop drastically when more than three sites are targeted simultaneously. The Synberc system combines Harvard’s Multiplex Automated Genome Engineering (MAGE) and Columbia’s Microarray Oligonucleotide (MO), to create MO-MAGE. MAGE allows simultaneous mutagenesis of multiple target sites in bacterial genomes using short oligonucleotides (such as the RNA or DNA molecules used in genetic research). However, this requires hundreds to thousands of unique oligos, which are costly to synthesize and impossible to scale up by traditional approaches. MO was developed to address the problem—it increases the number of oligos for direct use in altering thousands of genomic sites simultaneously.
One of the key milestones of synthetic biology is the development of “libraries” of diverse, well-characterized biological components that can be assembled to form new systems. The researchers’ vision for the MO-MAGE method is that it will be used to make thousands of specific chromosomal changes predicted to result in a desired phenotype (i.e., observable traits such height, eye color, and blood type) and combined with selection or screening of the cell library for promising mutants.